Abstract
The NF Data Portal community is dedicated towards bringing the latest data and research from the NF research community to the world. In doing so the Portal maintains and implements FAIR standards to make various types of research data findable, accessible, interoperable and reusable. Recently the Portal community reached out to the research community asking feedback regarding sharing logistics of various data types and their file formats. This document describes data types and formats that should be shared by the Neurofibromatosis (NF) research community for the NF Data Portal.
Introduction
The NF Data Portal is a collaborative data-sharing platform for Neurofibromatosis, a rare genetic disease (Allaway et al. 2019). It has been jointly supported by multiple funding organizations – the Gilbert Family Foundation (GFF), Children’s Tumor Foundation (CTF), Neurofibromatosis Therapeutic Acceleration Program (NTAP), NCI Developmental and Hyperactive Ras Tumor SPORE (DHART SPORE), CDMRP Neurofibromatosis Research Program (CDMRP NFRP), and Neurofibromatosis Research Initiative (NFRI) – and therefore represents substantial NF research activity.
As the NF Data Portal community continues to expand to add newly affiliated as well as independent researchers, and therefore new data, having these standards will help maintain the quality and quantity of our valuable resources. But also important is that this document should actually benefit our data contributors. One advantage is allowing our contributors to prioritize submissions accordingly. As well, our contributors have a clear reference for the data formats that are preferred for sharing. These specifications should lead to higher-impact data through encouraging interoperable formats that make data more likely to be reused (and cited). Finally, this open document lets our community be informed of the current state of art, including specifications currently not included, so that they can provide input towards specific/new data formats.
The data-sharing specifications highlighted in this document have been informed by members in the research community, using their opinions solicited through an open Request for Comments (RFC). The feedback received from the community was especially influential for data types outside of what is required by default, i.e. more “optional” data types. These specifications also follow requirements already established by the NF-OSI funding organizations, which apply to data types with clear “reusability” value and for which there are well-defined strategies for sharing (e.g. sequencing data). Journal requirements and other repositories were considered to a much lesser extent.
Data types and formats summary
As a representation of the most recent funder requirements and RFC results, table 1 summarizes specifications for common data types that are of interest for the NF community. These standards should apply for most general cases. However, we do note that for some data types, the community provided additional considerations that serve to qualify or supplement what is in this table (see Additional considerations). This qualitative feedback point to certain data domains that may need deeper evaluation. Alongside the qualitative results, quantitative breakdowns for components of the RFC are available (see RFC statistics).
| Requirement | Levelsa | Format | Notes | |
|---|---|---|---|---|
| DNA | ||||
| whole genome sequencing | required | raw OR semi-processed |
raw: FASTQ, unaligned BAM, CRAM | semi-processed: aligned BAM | |
| whole exome sequencing | required | raw OR semi-processed |
raw: FASTQ, unaligned BAM, CRAM | semi-processed: aligned BAM | |
| SNP microarray | required | raw AND processed |
raw: CEL, IDAT, tsv (raw values per SNP) & processed: tsv (genotypes per SNP) | |
| immunosequencing | required | raw OR semi-processed |
vendor-dependent, e.g. ImmunoSEQ and 10XGenomics formats | |
| Sanger sequencing | optional | processed |
||
| RNA expression | ||||
| RNA sequencing (bulk or single-cell) | required | raw OR semi-processed |
raw: FASTQ, unaligned BAM, CRAM | semi-processed: aligned BAM | |
| gene expression microarray | required | raw AND processed |
raw: CEL, IDAT, tsv (raw values per SNP, copy number, and loss of heterozygosity) & processed: tsv (normalized values and purity/ploidy) | |
| qPCR | optional | processed |
csv/tsv (according to template) | |
| methylation | ||||
| ATAC sequencing | required | raw OR semi-processed |
raw: FASTQ, unaligned BAM, CRAM | semi-processed: aligned BAM | |
| methylation array | required | raw OR semi-processed |
raw: FASTQ, unaligned BAM, CRAM | semi-processed: aligned BAM | |
| bisulfite sequencing | required | raw OR semi-processed |
raw: FASTQ, unaligned BAM, CRAM | semi-processed: aligned BAM | |
| protein | ||||
| LC-MS | required | raw AND processed |
raw: mzML & processed: protein intensities (csv/tsv) | https://www.psidev.info/mzML |
| western blot | optional | processed |
densitometry output (csv/tsv) | |
| plate-based ELISA | optional | raw |
plate reader output (csv/tsv) | |
| protein/peptide microarrays | required | processed |
label-free quantification matrix (csv/tsv) | |
| metabolomics | ||||
| LC-MS | required | raw AND processed |
raw: mzML or vendor-dependent format & processed: metabolite intensities (csv/tsv) | |
| clinical | ||||
| structured clinical data | required | processed |
csv/tsv or XML with metadata for each variable | key primary and secondary endpoints only |
| EEG | required | raw |
pending additional comments | |
| clinical/imaging | ||||
| MRI or other radiological image | required | raw |
dicom, nifti, minc | |
| imaging | ||||
| immunohistochemistry | required | raw |
a bio-formats compatible file format | https://www.tinyurl.com/bio-formats |
| immunofluorescence | required | raw |
a bio-formats compatible file format | https://www.tinyurl.com/bio-formats |
| gross morphology photos (mice) | optional | raw |
tiff, png, jpg or a bio-formats compatible file format | https://www.tinyurl.com/bio-formats |
| in vitro drug screening | ||||
| plate-based cell viability assay | required | processed |
csv/tsv (according to template) | |
| other | ||||
| flow cytometry | optional | raw |
fsc with gating parameters | |
| in vivo tumor growth experiments | optional | raw OR processed |
csv/tsv (according to template) where raw: tumor dimensions or other raw measurements & processed: calculated tumor volume/size | |
| a Level nomenclature can be cross-referenced with https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/data-levels, where 'raw' corresponses to Level 1 and 'semi-processed' most closely corresponds to Level 2. | ||||
Additional considerations
The current specifications should guide contributions in the “spirit of effective data sharing” and are most applicable when they lead to intended results, e.g. higher-impact shared data and reasonable balance of contributor/administrative effort given the expected value of the data shared. But there can be debatable cases where a “required” specification is less applicable. The community has anticipated issues like these and provided recommendations for some of them. However, difficult and unclear cases remain or may yet emerge, which will need more community consensus still.
Imaging data
Comments from the community noted two possible issues for imaging data.
A first question is whether imaging data should be shared when there are at most only a few images, which is of limited utility for re-analysis (e.g. with machine learning) usually requiring a large set of images. That is, imaging data may require a “critical mass”. When only a few images are contributed, these are likely only representative images that might also appear in a publication. In this case, it may not be really “required” that these images have to be shared.
A second case is for a study that may generate many images but can only process and fully annotate a subset. For example, there can be a large number of pathology images without any annotations of disease features by a pathologist, as these kinds of annotations are manually intensive. If all data are expected to be annotated – and annotations are especially important for the usefulness of the images – the question arises whether images outside the fully annotated subset should be shared. A clear suggestion seems to be that sharing images without annotations is better than not sharing images – for some images, it is possible for someone else to go back and add annotations. Note that basic, automatic annotations are still expected; using the example above, the images should still have an associated sample ID and tissue source.
Clinical data
Clinical data can be extremely diverse and unstructured. While we have incorporated comments for structured clinical data, we have not received comments for unstructured clinical data such as patient histories that are often captured in text reports. Clinical data still remains very open to comment from those in the community.
Alternative assay/platform data
The specifications table includes the most common assay data types instead of enumerating all variants or alternatives. An insightful community observation suggested there can be less common but better alternatives for some assays. The example given was using a WES capillary protein analysis system in lieu of traditional western blot for easier analysis as well as raw data sharing.
This is actually asking a different design question – not necessarily “What level and format of data should be used for better sharing?” but “What assays/platform should be used for better sharing?” It is certainly not within our purview to specify what should be used, and experiments will have to be planned around what assays the researchers are familiar with and what equipment is available. We are also aware that researchers are not likely to think about data sharing specifications until results have been generated, while this kind of consideration would take place before experiments even start. Nevertheless, if the investigator is at the juncture of a design decision, awareness of alternatives and their relative merit in terms of data sharing could be helpful. Unfortunately, this aspect is not captured well in our current specifications. We welcome the community providing more suggestions in this regard.
RFC statistics
Respondent profiles
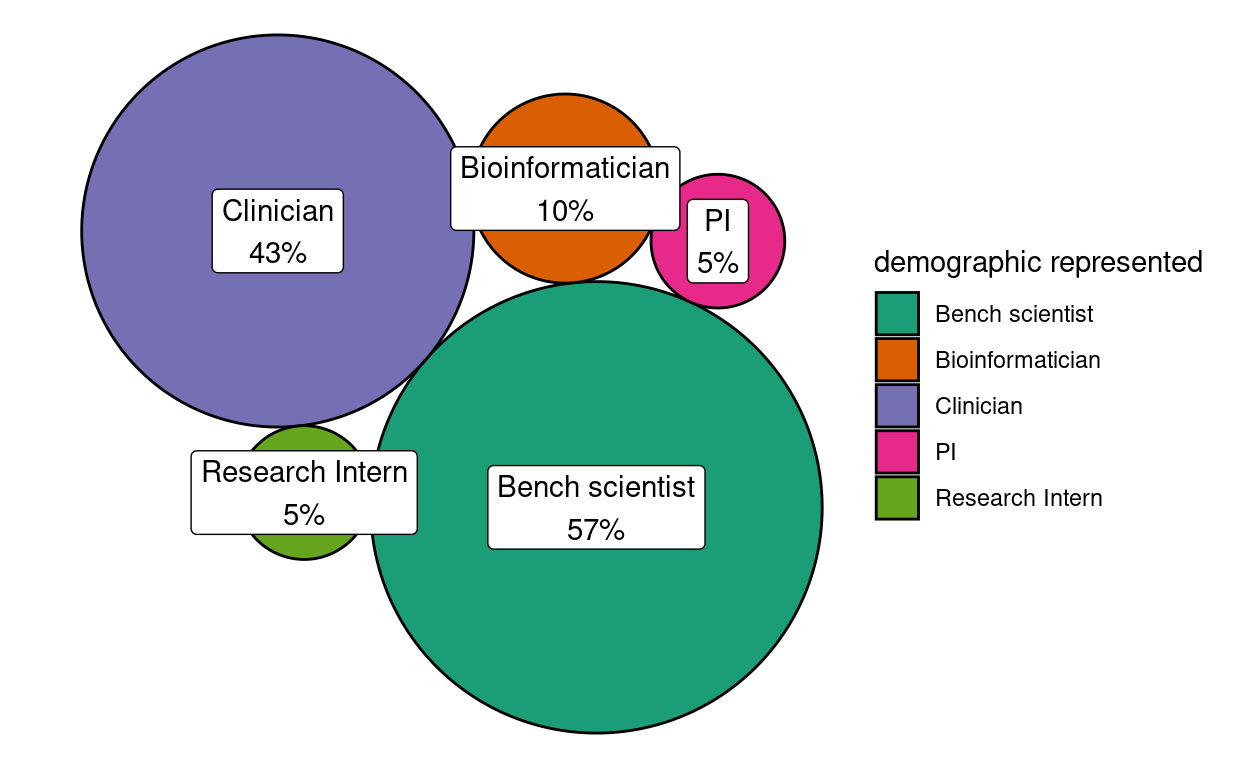
Figure 1: Demographic representation of RFC community respondents
Figure 1 summarizes the representation of different demographic types as self-reported through the question “What ‘type’ of NF research community member are you?” Because respondents could indicate more than one demographic type, a statistic of 43% here should be interpreted to mean that 43% of the responses represented the clinician perspective. Overall, the RFC garnered highest representation from bench scientists and clinicians, which was expected. That clinicians had nearly as much representation as bench scientists likely reflects the highly translational focus of the NF Data Portal community relative to more general data repositories.
In the RFC, we did not distinguish whether respondents considered themselves more a “data contributor”, a “data re-user” or possibly “equally both”. In the future, this additional facet may provide additional insight in case there are conflicts in perspectives between these profiles.
Survey quantified responses
Figure 2 summarizes responses to the question “Should it be required to deposit raw data?” for the different assay or different type categories. There were four categories that reached majority consensus for sharing: in vivo growth tumor data, Sanger sequencing, structured clinical data, and western blot.
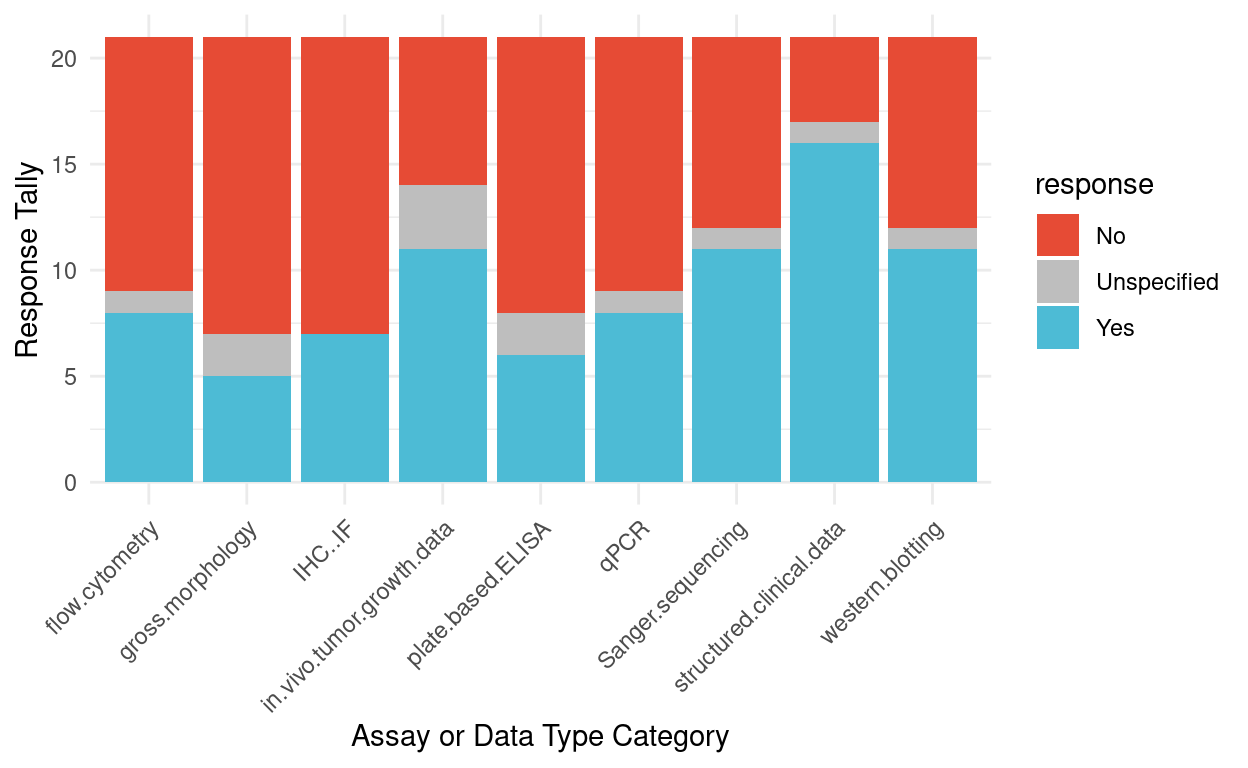
Figure 2: Survey responses for whether depositing raw data should be required for different assays
Figure 3 summarizes responses to the question “Have you ever re-analyzed this type of data, or wanted to, if the right dataset existed?” for the different assay or different type categories. The highest interest was for structured clinical data, which also garnered the strongest consensus that it should be shared (Fig 2).
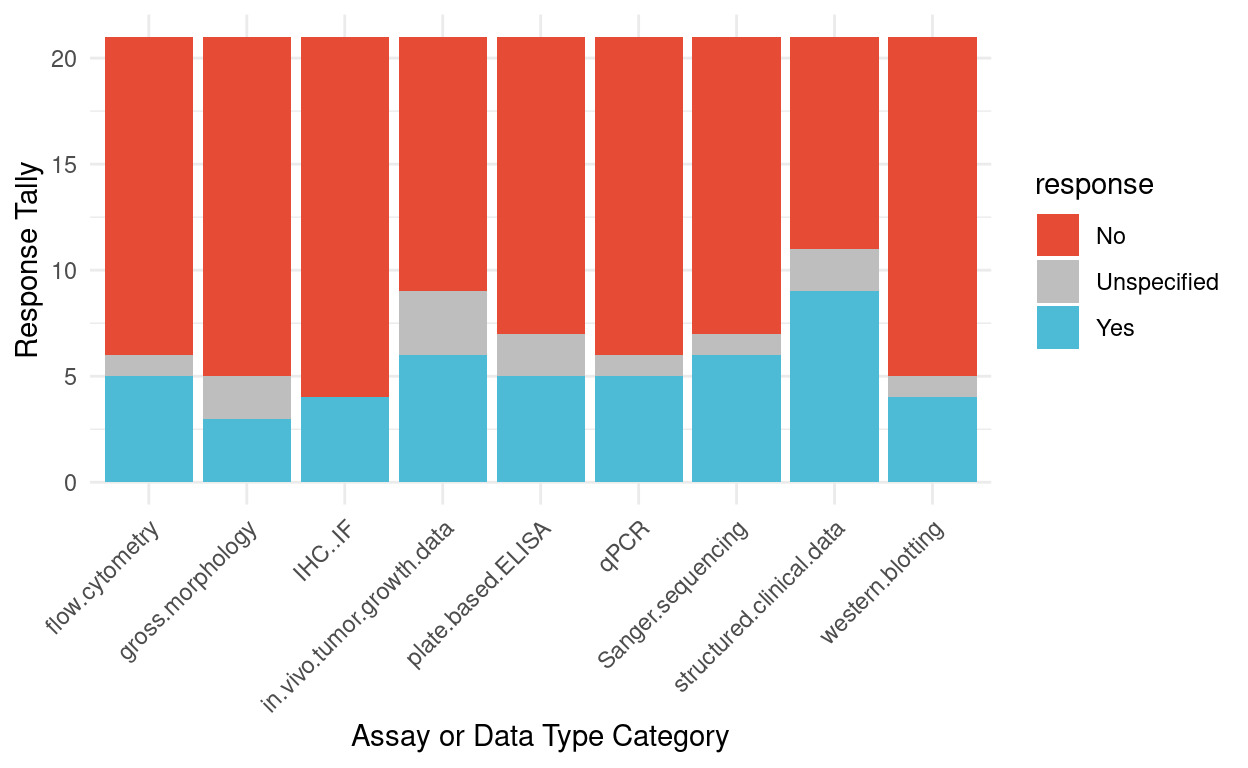
Figure 3: Survey responses for previous experience or interest in re-analyzing data for different assays
Figure 4 summarizes responses to the question “Have you previously been required to share a complete raw dataset by a funder or journal?” The responses suggest that funder or journal sharing requirements for these assays or data types are not common, except for perhaps structured clinical data and western blots. Compared to ratings in Fig 2, the results here also suggest that the community is willing to go beyond the “baseline” requirements of funders or journals.
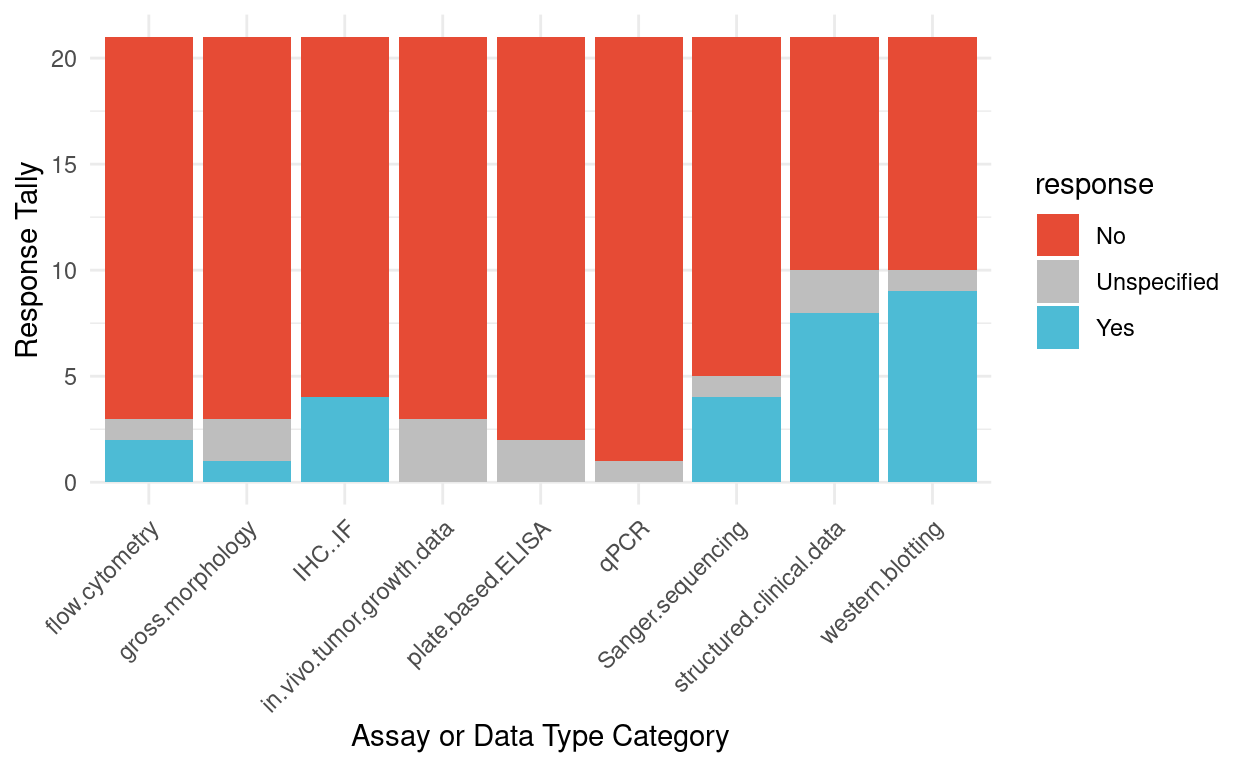
Figure 4: Survey responses for previous requirements to share raw data by a funder or journal